
eDNA—DNA that has been shed into the environment—tells the story of an ecosystem, the genetic signatures of plants, animals, fungi, microbes in a milieu that can be ‘snooped on’ by researchers as a non-invasive survey technique to do everything from detecting exotic species and contaminated waterways to informing palaeoecology and marine conservation. Photo: Santiago Ron/Flickr (Imbabura treefrog, Ecuador)
Act one: setting the stakes of an imperilled Earth
You know the story: the end is nigh and the fate of the entire world rests on the shoulders of the humble peasant, the apathetic nobody. Whether it’s Luke Skywalker on his desert planet polishing droids, or Neo pirating movies while robots threaten to mentally imprison all of humanity inside a fake world (I’m looking at you, Zuckerberg!), there are many stories where a lot rests on a little. And, tragically, it couldn’t be more true for the real world. At its time of need, the fate of our once pristine, now tattered natural world has been placed in the clumsy hands of an unlikely ape. What a twist: it’s us!
To rise up to this vital—perhaps most vital—call to arms, we humans (human as we are), developed the science of conservation biology. Conservation biology is about keeping Earth’s habitats and biodiversity alive and thriving, resplendent in its variation and precious in its instance nowhere but here on this Sun-smitten rock. This science is underpinned by a simple question: what exactly are we conserving? You need to know what’s out there to know whether it’s becoming more or less plentiful, and that means identifying it, counting it, and logging it. Loggerhead turtle? Check. Southern Bluefin tuna? Hmm…kinda. Dodo? Dodo!?
Identifying and counting individuals isn’t just fundamental for biodiversity’s sake, but also managing humanity’s appetite. Picture a fishing village that pegs their survival on a single lake of fish. If the village were naïve to the fish’s abundance, their movement, their demographics (which individuals are nearing reproductive age and must be returned), then fishing blind could spell starvation for all. In fact, how do you even count fish? They’re underwater! Sheep? Easy. Crows? Done. Dracula, even. But fish!?

Fish aren’t the only tricky ones to count: prawns, algae, bees, snow leopards, the tiny, the ephemeral, the indistinguishable and indistinct, all are challenges to an accurate snapshot. Photo: Michael Figiel/Flickr
Earth’s unlikely hero
As a species humans are not equipped to save the environment. Ecosystems, and the biodiversity they contain, operate at scales of complexity that defy our mortal senses: infrared, ultrasonic, microscopic, incomprehensibly large forest for the trees. It’s Animalia meets Pieter Bruegel the Elder in 3D. Our primate senses evolved to spot the tangible—ripe fruit, a knowing wink, the distant rumble of a savannah cat—not the odd versus even overlay of sub-millimetre scales that differentiate two species of fish larvae, one critically endangered, one hyper abundant.
On top of all that, it’s impossible to observe an ecosystem impartially. Human beings are smelly, noisy, large animals (technically ‘megafauna’, or ‘charismatic megafauna’ [but that depends who you’re speaking to]), so by entering an environment to measure it, we’re causing its observable biodiversity to shrink away. An ecologist in the field describing animals is, for all intents and purposes, ‘Fee-fi-fo-fum; I can’t smell the identity of any species in the vicinity’. To get an accurate census of an ecosystem, scientists have to deploy traps and snaps, and take samples with pricks and tricks to overcome our lumbering selves, to monitor the Schrodinger’s cat that is life on Earth.
And finally, regrettably, as Bono says, we’re ‘stuck in a moment and we can’t get out of it’. Ecologists literally sit and watch and listen and count up animals, a task only possible when everyone is there at the same time. There are clues, like a footprint or a scat (skibblee doowop), but otherwise, we’re captives in the present, our senses useful in this frame of life just now. This isn’t the only way. Take the nose of a dog—entering a room the dog ‘sees’ the ecosystem through time, the groups of people and activities happening last night, the toilet manner of others past and their identity. Wrong saviour species to choose, Earth.

Yes, dogs can smell through time—useful not just for sleuthing the past, but also establishing dominance over territory into the future. The identifying tag of pee makes for a more potent and idiosyncratic sign than any visual graffiti a youth might spray. Photo: Frank Shepherd/Flickr
Things are looking pretty dire for Earth: we’re in the midst of act two, we’ve got plenty of tools and techniques for cracking conservation, but as a science-conducting species we’re crippled by mediocrity. All of this means…
We’re gonna need a montage (montage)
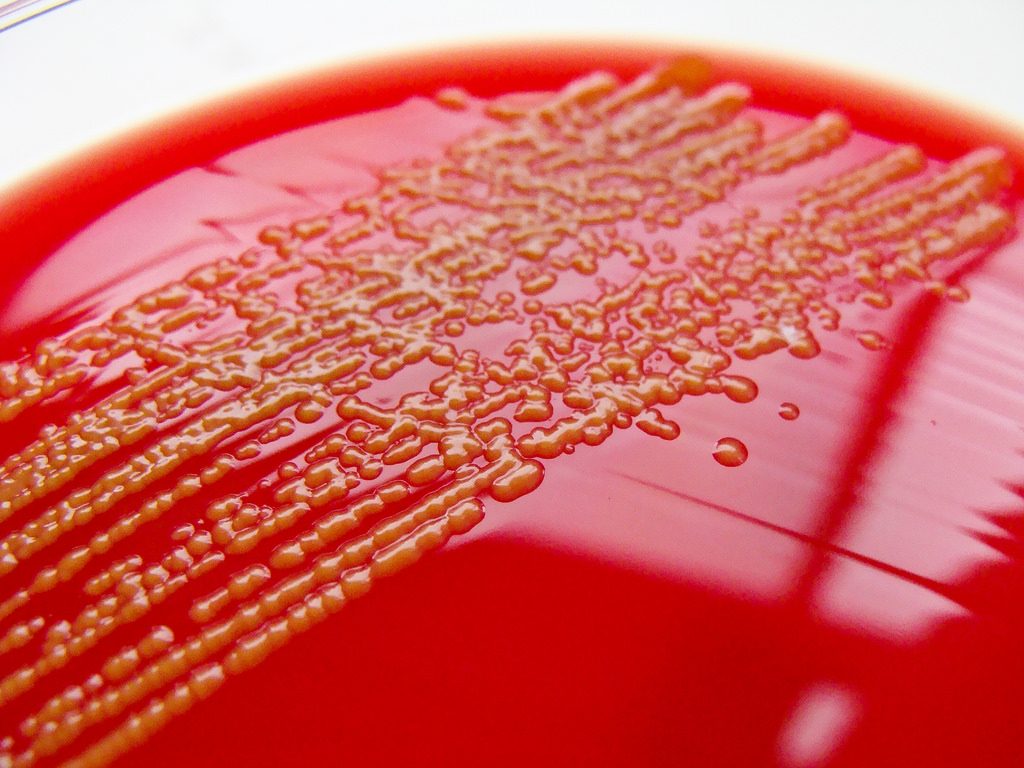
Life is messy; difficult to cultivate, to study, to understand. Photo: Nathan Reading/Flickr (S. paucimobilis: a slow-moving soil-based bacterium associated with various clinical infections.)
Meanwhile, microbiologists were facing the following problem with their tiny subjects: to identify a species of microbe it must be grown in culture, but only some species can grow in culture. This means—just like an ecologist in a jungle—they were cataloguing only a portion, a subset of biodiversity in a given microbial ecosystem. So what’d they do?
With technological advancements burgeoning (you go, science!), they thought: Why bother growing a creature to identify it directly, when you can do it indirectly using the clue that betrays it the most: DNA!
By extracting the DNA from the microbes’ cells, called ‘intracellular’ DNA, researchers were able to identify the species of a sample, not just that, but given a complex sample of many species and other gunk they can simultaneously identify all the species contained therein. (So long as they have a DNA sequence catalogue to compare against.) All this is possible given the inherent differences between species and their uniquely defined DNA sequences, and at a finer level still—individuals, which can be distinguished from specific sections of their DNA.
Here comes the neat part: owing to DNA’s structural integrity, it doesn’t need to be inside the cells of a living creature for us to extract and analyse it. That’s right—it can just be floating around in the environment, so-called ‘extracellular’ DNA. We’re not going to patronise you by breaking the components of those DNA words—intracellular and extracellular—down, though you know what will? Everything else.
Yep, there’s plenty of DNA in the environment, like ghosts lingering over the old haunts of the creatures that shed it, the world where they lived. Take us for example, the aforementioned damn dirty apes. Every hour you shed around a million particles into the air, a ‘microbial cloud’ that’s uniquely yours: oil smears at every turn, hair ejections, skin cells like tiles in a typhoon. Twenty four hours in a room and it’s basically you. CSI can nail you sometime after fleeing the scene of a crime, or even, a misfire at the bowl. This goes for every other organism, too: fish piss and loose feathers, fallen leaves and fungal spores, coral spawn and blood in the water—all of it revelatory, a window to an ecosystem.
DNA doesn’t just hang around waiting to be analysed, though. As robust as the double helix molecule is, it’s also a valuable resource and one that gets broken down by UV rays, chemicals, life, too. DNA’s built on twin backbones of sugar and phosphate, zipper teeth of nitrogen. All nutritious all the time. Sound delicious? You’re not alone. Free-floating DNA in an ecosystem is like Fiddy Cent’s ham-fisted Candy Shop metaphor, with microbes and fungi only too happy to ‘break it down for you now; baby it’s simple [monomers]’.
Though these fairy lights aren’t edible*, you know what is? DNA. That’s right, if you’re eating food, be it plant or animal products, you’re also eating whole DNA or components, in-turn used to build your own genetic make-up. Keep going till you hit the spot, woah. Photo: Stuart Caie/Flickr (*We couldn’t find a nice food-based DNA picture.)
And not only is DNA getting eaten by things but it’s washed away, too: fish excreta in the ocean, owl effluent in a river, all that evidence lost in time, like cat spray in rain…
How does it work? Turn up the PCR, blast your DNA, pump it (louder)
As any eDNA-wielding ecologist will tell you: before entering the field, it’s a good idea to have a focus of the study, the species, the population you’re analysing. Then, you get a sample. This can be as broad as it sounds: a beaker of seawater from Ningaloo, melt water from a two million year old ice core, some sodden soil in the lion enclosure of a zoo—all examples from actual eDNA research studies.
Samples, though, are not so simple; as mentioned, DNA is on the move. If you’re sampling seawater, the currents, the salinity, the temperature, all will influence your sample. Hot and fast-moving currents mean your DNA will likely be young, since anything older will have moved on or broken up. Cold, slow-moving environments, on the other hand, mean a DNA trace that might stretch back millennia. You’ve got to know your environment and factor that in. This might mean preliminary, reiterated control trials before you get your feet wet.

Safety first. “Environomics will build new ways to see beyond the landscape to its underlying genescape. Photo: Center for International Forestry Research (CIFOR)”
Got your sample? Let’s extract the DNA. We won’t freak you out with the nitty gritty of PCR and DNA amplification—a process whose intricate ins and outs this blog author definitely, definitely understands—but just know that researchers have electric jelly that duplicates DNA by zapping it, a bit like Hugh Jackman’s twist bad guy in The Prestige (*spoiler alert soz 2late neway*).
There are basically two ways to use eDNA data. 1) Species-specific: If you know which species you’re looking for then you can rake through the sample’s DNA until you find a match, telling you if your creature is or isn’t there. 2) DNA metabarcoding: About as cool as it sounds. Imagine a robot doing a find-a-word. You need to give it the word puzzle, but also a dictionary to use as a database. Your environmental DNA sample is the puzzle, and a range of online, genome-level databases are the dictionary, and with the exploratory spirit, you can comb the environment, identifying which species are present. These databases can be patchy, sometimes only identifying broader groupings such as: there’s a bear in there, and a cherry as well, etc. Ideally, though, these catalogues would be biological, linked to reference specimens such as those kept in our own national research collections.
Invisible apes. Teleporting apes. Time travelling apes.
eDNA is a powerful technique. With it we can transcend our primate constraints—and, though new, already its applications have been vast and stunning. In fact, the first published paper to apply eDNA was a tough act to follow.
This team went to Greenland and took ice cores. These cores were kilometres deep, tunnelling through glaciers, icy silt, and earth, down through layers and back through time. And, like that scene from Jurassic Park—the shaft of the needle through the amber core, edging towards the mosquito—instead of insect chitin, these ice cores pierced the Earth’s crust, and the DNA they syringed, not that of an individual, but of an ancient ecosystem not exposed to warm air or animal eyes for over two million years, resurrected here for us now, uncovered were plants in the family of roses and strawberries, the DNA of butterflies and moths that would have flitted and showered pollen and scales.

Blood of an insect frozen in that of a tree. Clever girl. Photo: Graeme Churchard/Flickr
The research didn’t stop there. Ancient DNA from North America of long-extinct horses and mammoths that—abutting against fossil evidence—showed sustained peaceful co-existence with humans, eDNA being used to identify the diversity of animals of a zoo, ostrich farms in Denmark, the earthworm spread of ecosystems feeding up through aerated, nutrient-rich soil to the diversity above, and even the dark and icy waters of Loch Ness, searching for DNA evidence of the unknown, and, when the reference database isn’t sufficient to define enigmatic DNA sequences, using placeholder MOTUs—Molecular Operational Taxonomic Units—the equivalent of unidentified species silhouettes (if you’re from the 90s generation, Who’s That Pokémon!?).
CSIRO down…you’re moving too fast
Unfortunately we have to abandon this article’s extended metaphor about an imperilled Earth and an unlikely hero because science isn’t a narrative with a dénouement or a conclusion, only caveats and next steps.
The downsides of eDNA are plenty: we’re limited by catalogues, changeability of each ecosystem, persistence of eDNA—but perhaps more imposing are the upsides. For centuries, scientists have dealt only with the tangible, netting insects, decanting formaldehyde, penning labels, becoming intimate with these physical techniques, their short-comings and strengths, the methods and results, and how best to interpret them. Now with eDNA we’re disembodied from the visible. As with any new technology, they grant us ‘superhuman’ powers, setting up new ways of thinking, pathways for us to grow into and this is no exception. eDNA is a tool whose mastery promises results limited only by our ability to interpret them.
And this chasm, between the familiar and the novel, grows with each innovation. Just like iPhones falling off the production line with ever-glamorising ways to take a selfie (can we put down the stick now?), DNA-sequencing technology develops rapidly, presenting more applications, more results, more challenges. And it’s all very new. eDNA is an emerging technology, one that will find its place—with use—among the growing biomonitoring toolkit for both its utility and its short-comings.
Environomics
“Using genomics, bioinformatics and simulation, Environomics is finding new resources in nature and reinventing how we measure and monitor ecosystem health, change and threats.”
Environomics is the name of one of our Future Science Platforms, a multi-disciplinary team of researchers using eDNA among seven other projects, all with thrilling new applications. We’re measuring the biodiversity of fishes in marine ecosystems, working towards monitoring their population numbers for drops and warning signs, and pushing the boundaries of what eDNA can tell us: not just which species are there, but which specific individuals, their age, their genetic diversity.
eDNA can be wielded wherever a researcher’s imagination might reach, where answers are most desperate, from an ingredient-revealing sample of your soup, to a test in drinking water for the presence of pathogens, and beyond. Science and technology, like that of eDNA, have empowered people to reach beyond ourselves, rise to the call to arms of protecting and conserving our planet, our Sun-kissed spacecraft and sole bearer of life.


